Specifiyng weights in Log-rank comparisons
Marcin Kosinski
created 29-01-2017, revised 22-08-2018
Source:vignettes/Specifiying_weights_in_log-rank_comparisons.Rmd
Specifiying_weights_in_log-rank_comparisons.RmdThis vignette covers changes between versions 0.2.4 and 0.2.5 for specifiyng weights in the log-rank comparisons done in
ggsurvplot().
Log-rank statistic for 2 groups
As it is stated in the literature, the Log-rank test for comparing survival (estimates of survival curves) in 2 groups ( and ) is based on the below statistic
where and
- for are possible event times,
- is the overall risk set size on the time (),
- is the risk set size on the time in group ,
- is the risk set size on the time in group ,
- overall observed events in the time (),
- observed events in the time in group ,
- observed events in the time in group ,
- number of overall expected events in the time (),
- number of expected events in the time in group ,
- number of expected events in the time in group ,
- is a weight for the statistic,
also remember about few notes
that’s why we can substitute group with in and receive same results.
Weighted Log-rank extensions
Regular Log-rank comparison uses
but many modifications to that approach have been proposed. The most
popular modifications, called weighted Log-rank tests, are available in
?survMisc::comp
-
nGehan and Breslow proposed to use (this is also called generalized Wilcoxon), -
srqtNTharone and Ware proposed to use , -
S1Peto-Peto’s modified survival estimate , -
S2modified Peto-Peto (by Andersen) , -
FHFleming-Harrington .
Watch out for
FHas I submitted an info on survMisc repository where I think their mathematical notation is misleading for Fleming-Harrington.
Why are they useful?
The regular Log-rank test is sensitive to detect differences in late
survival times, where Gehan-Breslow and Tharone-Ware propositions might
be used if one is interested in early differences in survival times.
Peto-Peto modifications are also useful in early differences and are
more robust (than Tharone-Whare or Gehan-Breslow) for situations where
many observations are censored. The most flexible is Fleming-Harrington
method for weights, where high p indicates detecting early
differences and high q indicates detecting differences in
late survival times. But there is always an issue on how to detect
p and q.
Remember that test selection should be performed at the research design level! Not after looking in the dataset.
Plots
library("survival")
data("lung")
fit <- survfit(Surv(time, status) ~ sex, data = lung)After preparing a functionality for this GitHub’s issue Other tests than log-rank for testing survival curves and Log-rank test for trend we are now able to compute p-values for various Log-rank test in survminer package. Let as see below examples on executing all possible tests.
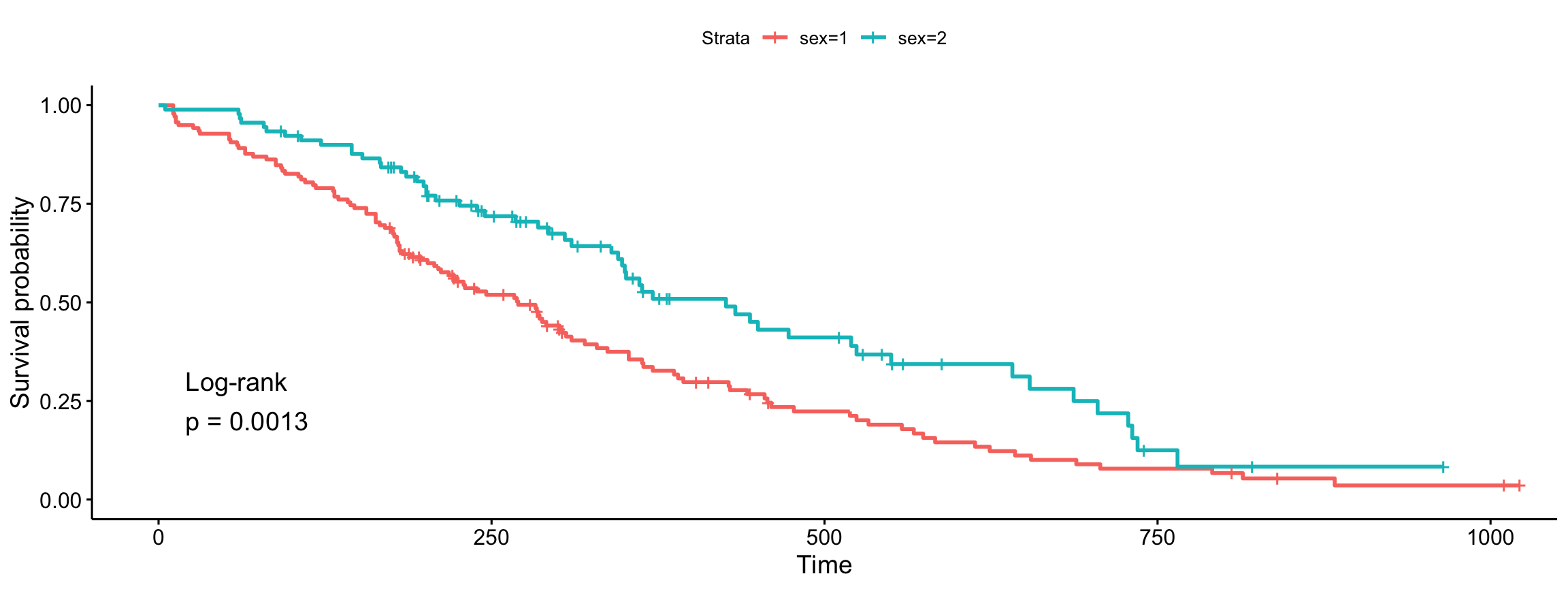
Log-rank (comp)
ggsurvplot(fit, data = lung, pval = TRUE, pval.method = TRUE,
log.rank.weights = "1")
Gehan-Breslow (generalized Wilcoxon)
ggsurvplot(fit, data = lung, pval = TRUE, pval.method = TRUE,
log.rank.weights = "n", pval.method.coord = c(5, 0.1),
pval.method.size = 3)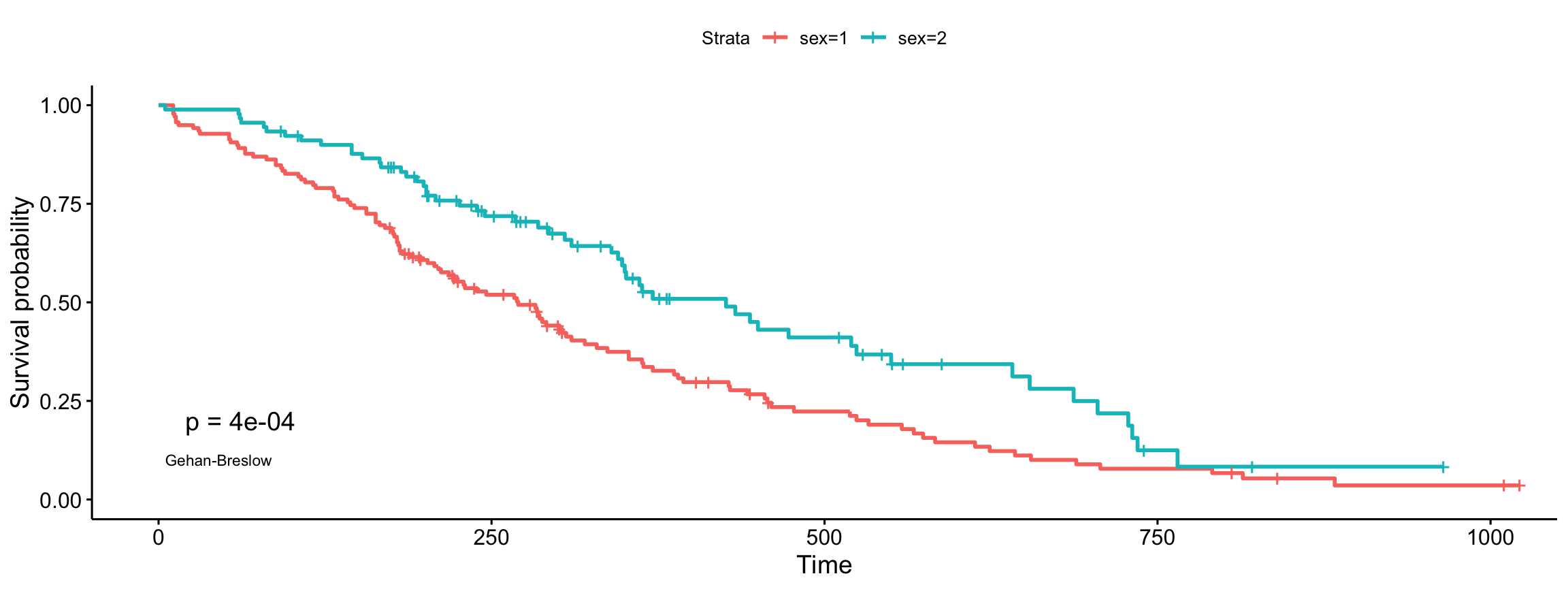
Tharone-Ware
ggsurvplot(fit, data = lung, pval = TRUE, pval.method = TRUE,
log.rank.weights = "sqrtN", pval.method.coord = c(3, 0.1),
pval.method.size = 4)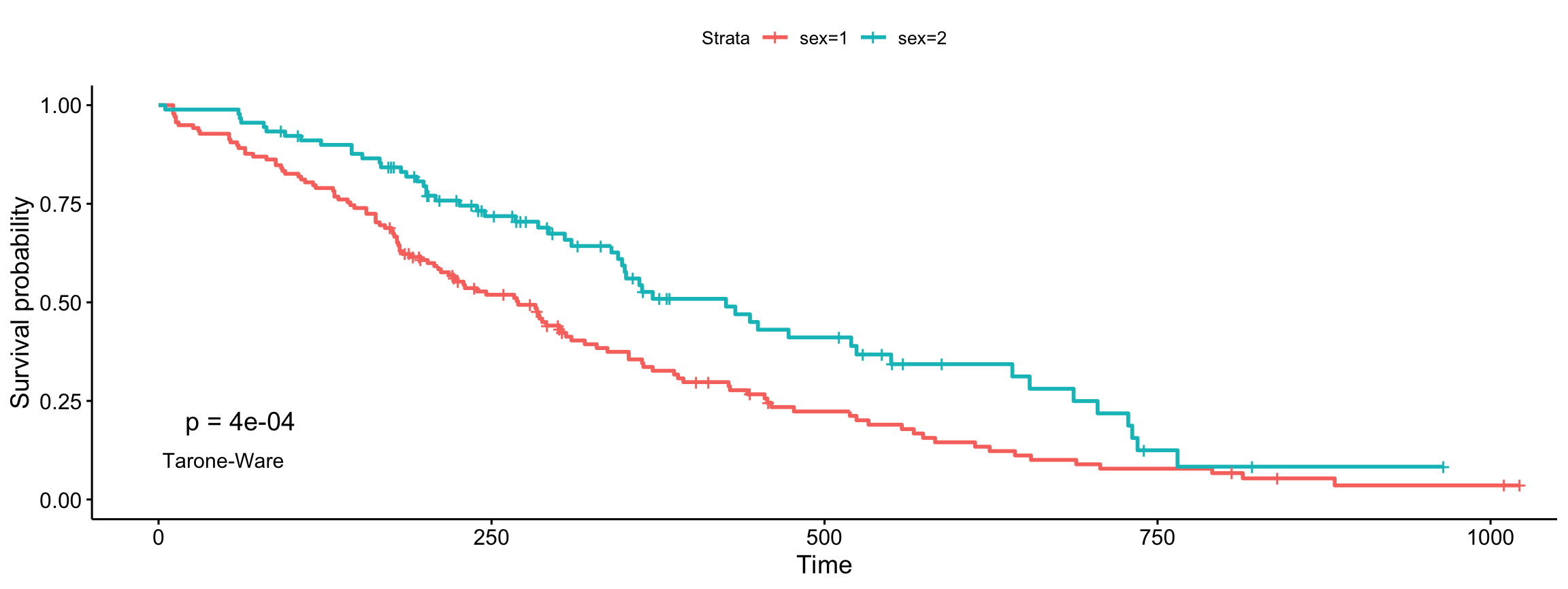
Peto-Peto’s modified survival estimate
ggsurvplot(fit, data = lung, pval = TRUE, pval.method = TRUE,
log.rank.weights = "S1", pval.method.coord = c(5, 0.1),
pval.method.size = 3)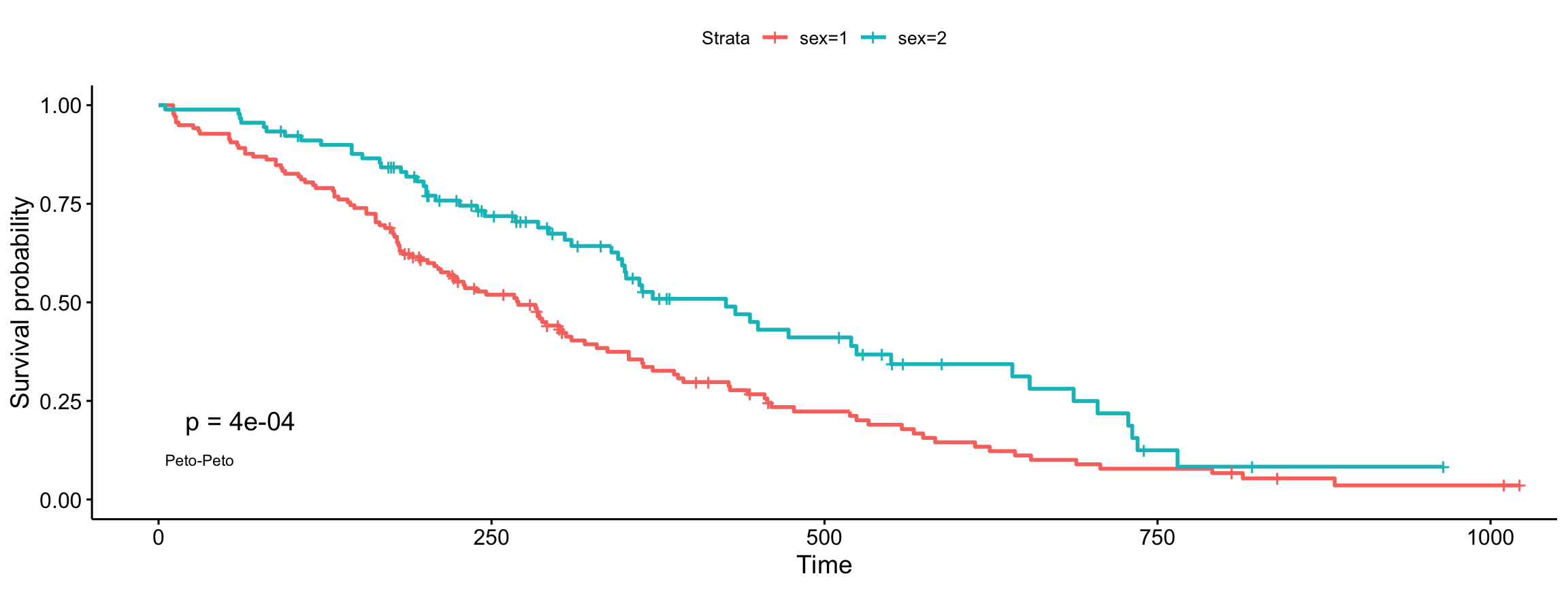
modified Peto-Peto’s (by Andersen)
ggsurvplot(fit, data = lung, pval = TRUE, pval.method = TRUE,
log.rank.weights = "S2", pval.method.coord = c(5, 0.1),
pval.method.size = 3)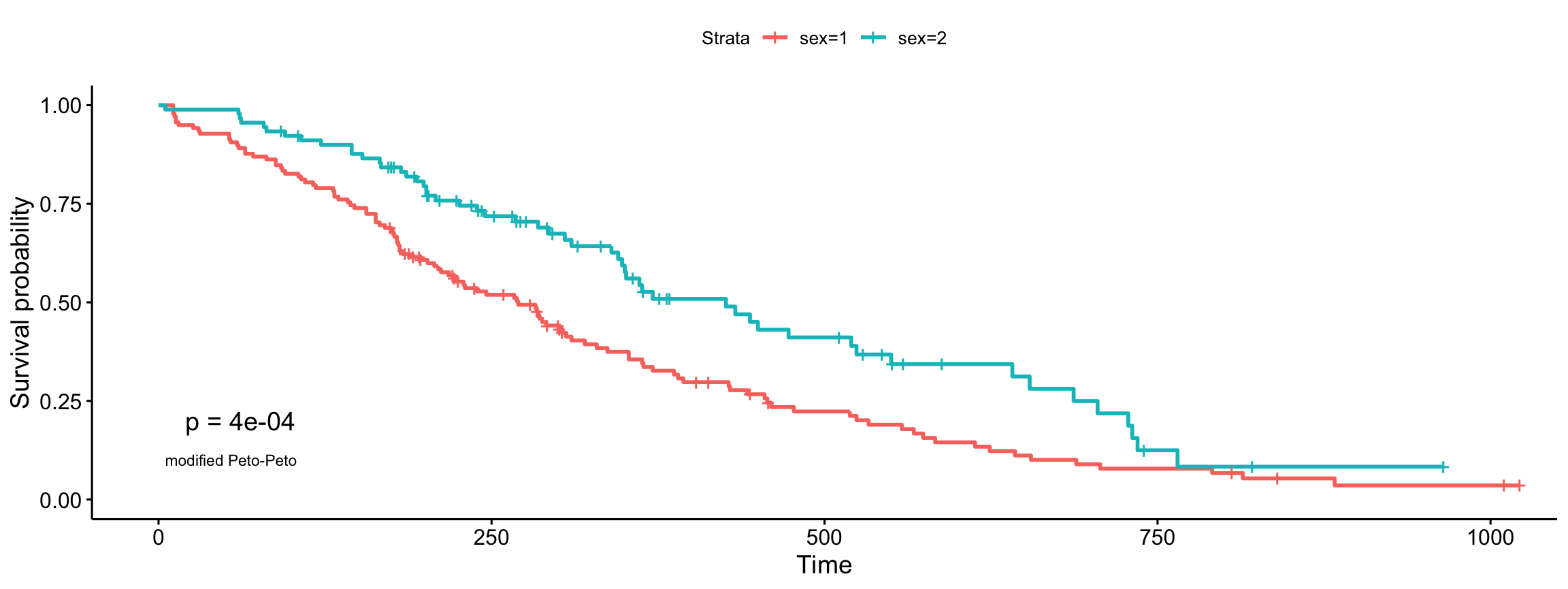
Fleming-Harrington (p=1, q=1)
ggsurvplot(fit, data = lung, pval = TRUE, pval.method = TRUE,
log.rank.weights = "FH_p=1_q=1",
pval.method.coord = c(5, 0.1),
pval.method.size = 4)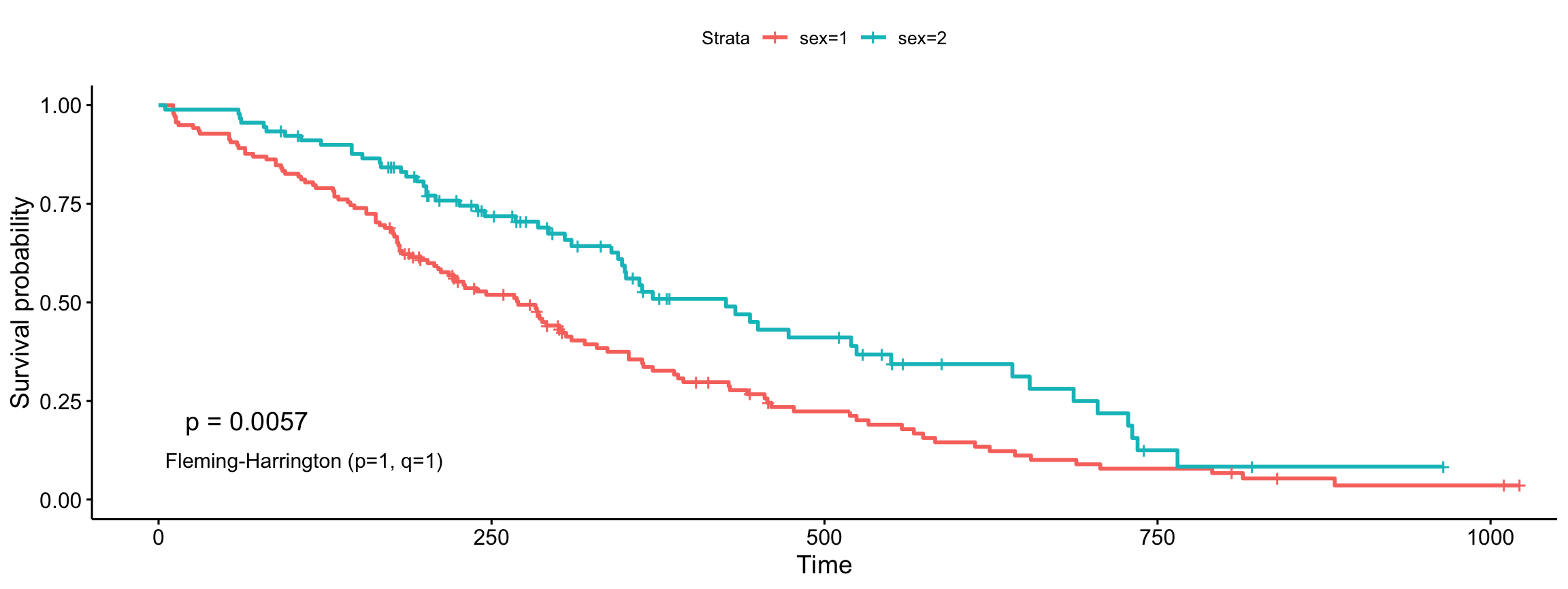
References
Gehan A. A Generalized Wilcoxon Test for Comparing Arbitrarily Singly-Censored Samples. Biometrika 1965 Jun. 52(1/2):203-23.
Tarone RE, Ware J 1977 On Distribution-Free Tests for Equality of Survival Distributions. Biometrika;64(1):156-60.
Peto R, Peto J 1972 Asymptotically Efficient Rank Invariant Test Procedures. J Royal Statistical Society 135(2):186-207.
Fleming TR, Harrington DP, O’Sullivan M 1987 Supremum Versions of the Log-Rank and Generalized Wilcoxon Statistics. J American Statistical Association 82(397):312-20.
Billingsly P 1999 Convergence of Probability Measures. New York: John Wiley & Sons.