Combine a List of Survfit Objects on the Same Plot
Source:R/ggsurvplot_combine.R
ggsurvplot_combine.RdCombine multiple survfit objects on the same plot. For example,
one might wish to plot progression free survival and overall survival on
the same graph (and also stratified by treatment assignment).
ggsurvplot_combine() provides an extension to the
ggsurvplot() function for doing that.
Usage
ggsurvplot_combine(
fit,
data,
risk.table = FALSE,
risk.table.pos = c("out", "in"),
cumevents = FALSE,
cumcensor = FALSE,
tables.col = "black",
tables.y.text = TRUE,
tables.y.text.col = TRUE,
ggtheme = theme_survminer(),
tables.theme = ggtheme,
keep.data = FALSE,
risk.table.y.text = tables.y.text,
...
)Arguments
- fit
a named list of survfit objects.
- data
the data frame used to compute survival curves.
- risk.table
Allowed values include:
TRUE or FALSE specifying whether to show or not the risk table. Default is FALSE.
"absolute" or "percentage". Shows the absolute number and the percentage of subjects at risk by time, respectively.
"abs_pct" to show both absolute number and percentage.
"nrisk_cumcensor" and "nrisk_cumevents". Show the number at risk and, the cumulative number of censoring and events, respectively.
- risk.table.pos
character vector specifying the risk table position. Allowed options are one of c("out", "in") indicating 'outside' or 'inside' the main plot, respectively. Default value is "out".
- cumevents
logical value specifying whether to show or not the table of the cumulative number of events. Default is FALSE.
- cumcensor
logical value specifying whether to show or not the table of the cumulative number of censoring. Default is FALSE.
- tables.col
color to be used for all tables under the main plot. Default value is "black". If you want to color by strata (i.e. groups), use tables.col = "strata".
- tables.y.text
logical. Default is TRUE. If FALSE, the y axis tick labels of tables will be hidden.
- tables.y.text.col
logical. Default value is FALSE. If TRUE, tables tick labels will be colored by strata.
- ggtheme
function, ggplot2 theme name. Default value is theme_survminer. Allowed values include ggplot2 official themes: see
theme.- tables.theme
function, ggplot2 theme name. Default value is theme_survminer. Allowed values include ggplot2 official themes: see
theme. Note that,tables.themeis incremental toggtheme.- keep.data
logical value specifying whether the plot data frame should be kept in the result. Setting these to FALSE (default) can give much smaller results and hence even save memory allocation time.
- risk.table.y.text
logical. Default is TRUE. If FALSE, risk table y axis tick labels will be hidden.
- ...
other arguments to pass to the
ggsurvplot()function.
Examples
library(survival)
# Create a demo data set
#::::::::::::::::::::::::::::::::::::::::::::::::::::::::
set.seed(123)
demo.data <- data.frame(
os.time = colon$time,
os.status = colon$status,
pfs.time = sample(colon$time),
pfs.status = colon$status,
sex = colon$sex, rx = colon$rx, adhere = colon$adhere
)
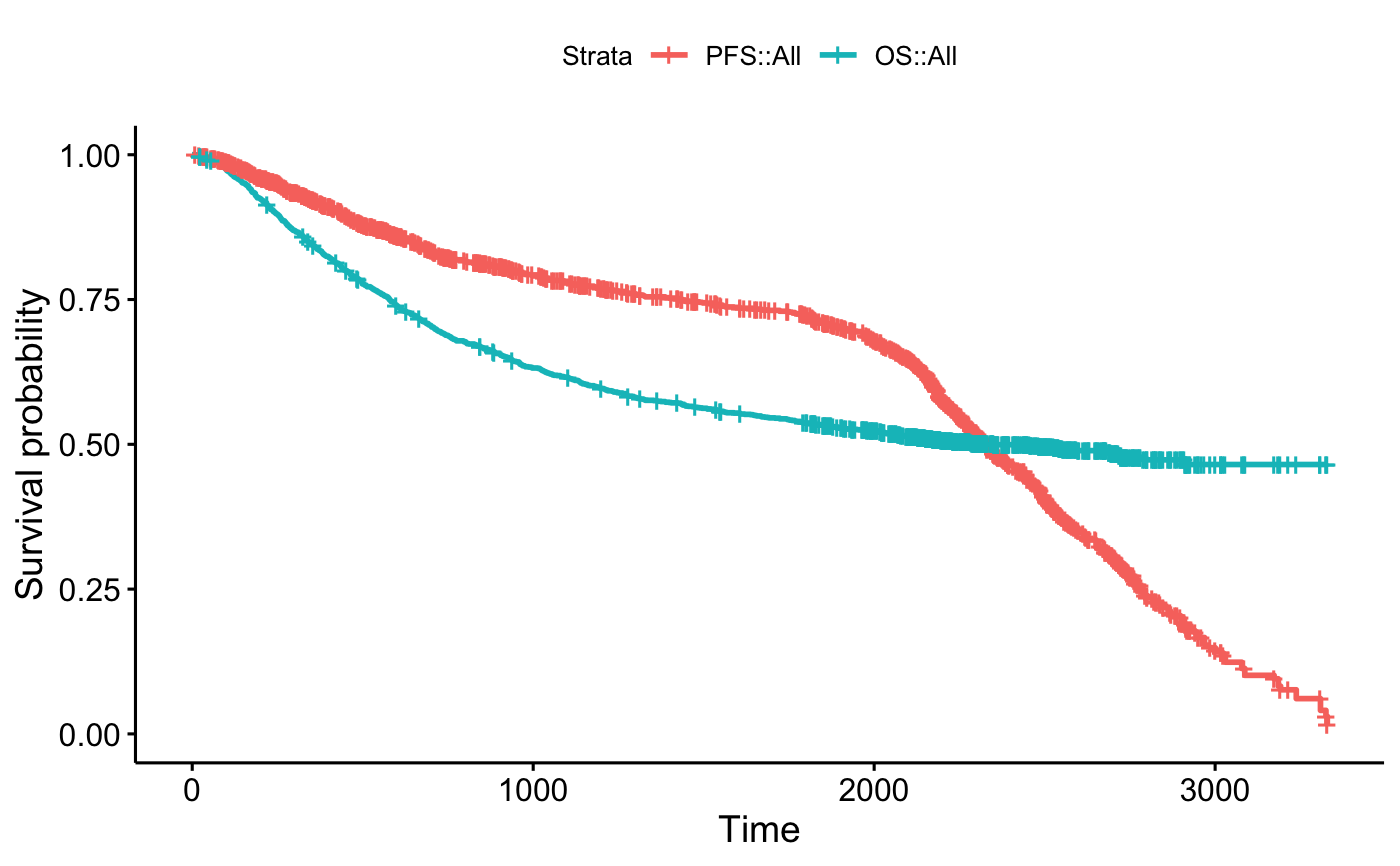
# Ex1: Combine null models
#::::::::::::::::::::::::::::::::::::::::::::::::::::::::
# Fit
pfs <- survfit( Surv(pfs.time, pfs.status) ~ 1, data = demo.data)
os <- survfit( Surv(os.time, os.status) ~ 1, data = demo.data)
# Combine on the same plot
fit <- list(PFS = pfs, OS = os)
ggsurvplot_combine(fit, demo.data)
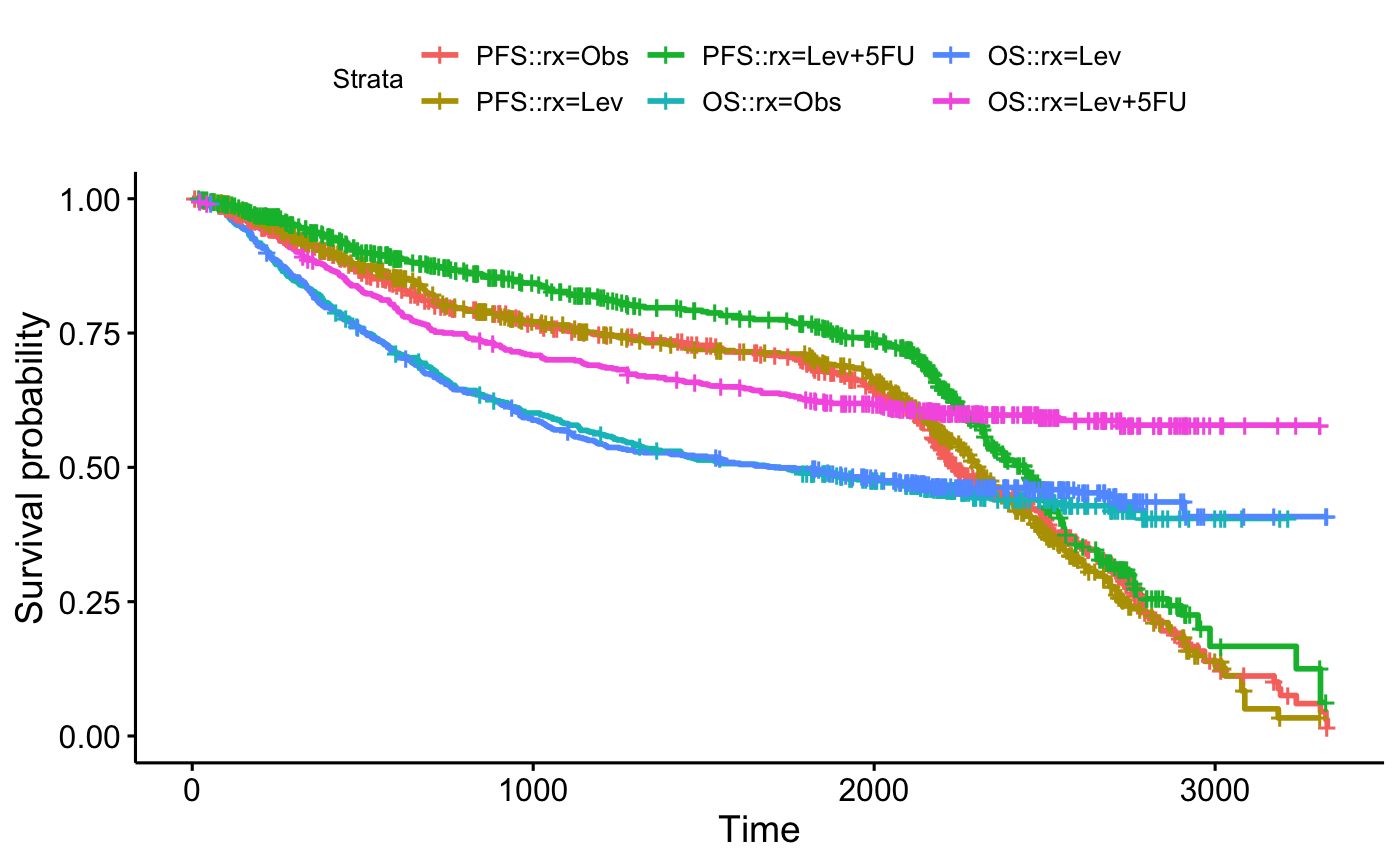
 # Combine survival curves stratified by treatment assignment rx
#::::::::::::::::::::::::::::::::::::::::::::::::::::::::
# Fit
pfs <- survfit( Surv(pfs.time, pfs.status) ~ rx, data = demo.data)
os <- survfit( Surv(os.time, os.status) ~ rx, data = demo.data)
# Combine on the same plot
fit <- list(PFS = pfs, OS = os)
ggsurvplot_combine(fit, demo.data)
# Combine survival curves stratified by treatment assignment rx
#::::::::::::::::::::::::::::::::::::::::::::::::::::::::
# Fit
pfs <- survfit( Surv(pfs.time, pfs.status) ~ rx, data = demo.data)
os <- survfit( Surv(os.time, os.status) ~ rx, data = demo.data)
# Combine on the same plot
fit <- list(PFS = pfs, OS = os)
ggsurvplot_combine(fit, demo.data)