Breat and Ovarian cancers survival information from the RTCGA.clinical R/Bioconductor package.http://rtcga.github.io/RTCGA/.
Usage
data("BRCAOV.survInfo")Format
A data frame with 1674 rows and 4 columns.
- times: follow-up time;
- bcr_patient_barcode: Patient bar code;
- patient.vital_status = survival status. 0 = alive, 1 = dead;
- admin.disease_code: disease code. brca = breast cancer, ov = ovarian
cancer.
Examples
data(BRCAOV.survInfo)
library(survival)
#>
#> Attaching package: ‘survival’
#> The following object is masked from ‘package:survminer’:
#>
#> myeloma
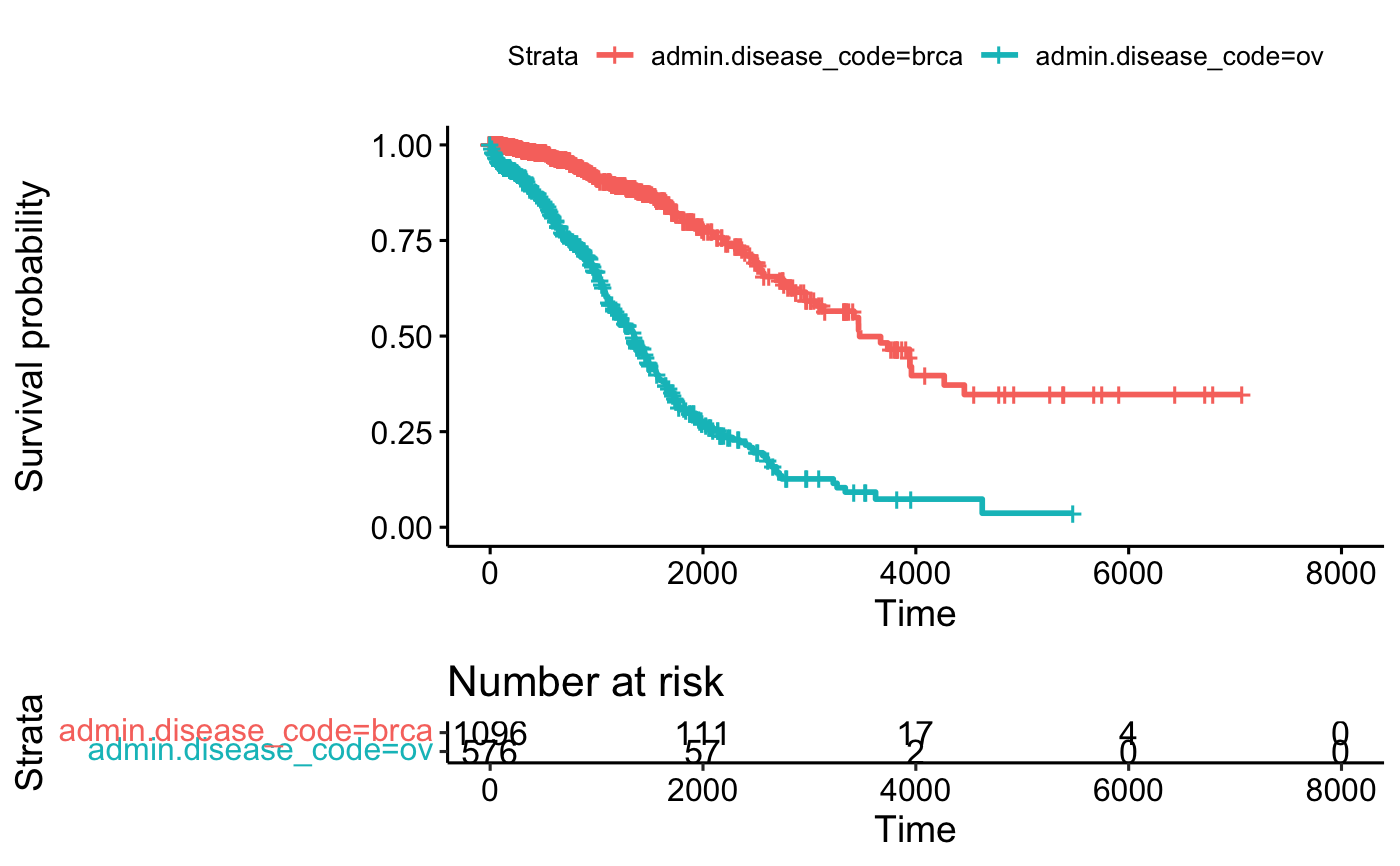
fit <- survfit(Surv(times, patient.vital_status) ~ admin.disease_code,
data = BRCAOV.survInfo)
ggsurvplot(fit, data = BRCAOV.survInfo, risk.table = TRUE)
#> Warning: Using `size` aesthetic for lines was deprecated in ggplot2 3.4.0.
#> ℹ Please use `linewidth` instead.
#> ℹ The deprecated feature was likely used in the ggpubr package.
#> Please report the issue at <https://github.com/kassambara/ggpubr/issues>.
#> Ignoring unknown labels:
#> • colour : "Strata"