Visualize the contributions of row/column elements
fviz_contrib.RdThis function can be used to visualize the contribution of rows/columns from the results of Principal Component Analysis (PCA), Correspondence Analysis (CA), Multiple Correspondence Analysis (MCA), Factor Analysis of Mixed Data (FAMD), and Multiple Factor Analysis (MFA) functions.
fviz_contrib(X, choice = c("row", "col", "var", "ind", "quanti.var", "quali.var", "group", "partial.axes"), axes = 1, fill = "steelblue", color = "steelblue", sort.val = c("desc", "asc", "none"), top = Inf, xtickslab.rt = 45, ggtheme = theme_minimal(), ...) fviz_pca_contrib(X, choice = c("var", "ind"), axes = 1, fill = "steelblue", color = "steelblue", sortcontrib = c("desc", "asc", "none"), top = Inf, ...)
Arguments
| X | an object of class PCA, CA, MCA, FAMD, MFA and HMFA [FactoMineR]; prcomp and princomp [stats]; dudi, pca, coa and acm [ade4]; ca [ca package]. |
|---|---|
| choice | allowed values are "row" and "col" for CA; "var" and "ind" for PCA or MCA; "var", "ind", "quanti.var", "quali.var" and "group" for FAMD, MFA and HMFA. |
| axes | a numeric vector specifying the dimension(s) of interest. |
| fill | a fill color for the bar plot. |
| color | an outline color for the bar plot. |
| sort.val | a string specifying whether the value should be sorted. Allowed values are "none" (no sorting), "asc" (for ascending) or "desc" (for descending). |
| top | a numeric value specifying the number of top elements to be shown. |
| xtickslab.rt | Same as x.text.angle and y.text.angle, respectively. Will be deprecated in the near future. |
| ggtheme | function, ggplot2 theme name. Default value is theme_pubr(). Allowed values include ggplot2 official themes: theme_gray(), theme_bw(), theme_minimal(), theme_classic(), theme_void(), .... |
| ... | other arguments to be passed to the function ggpar. |
| sortcontrib | see the argument sort.val |
Value
a ggplot2 plot
Details
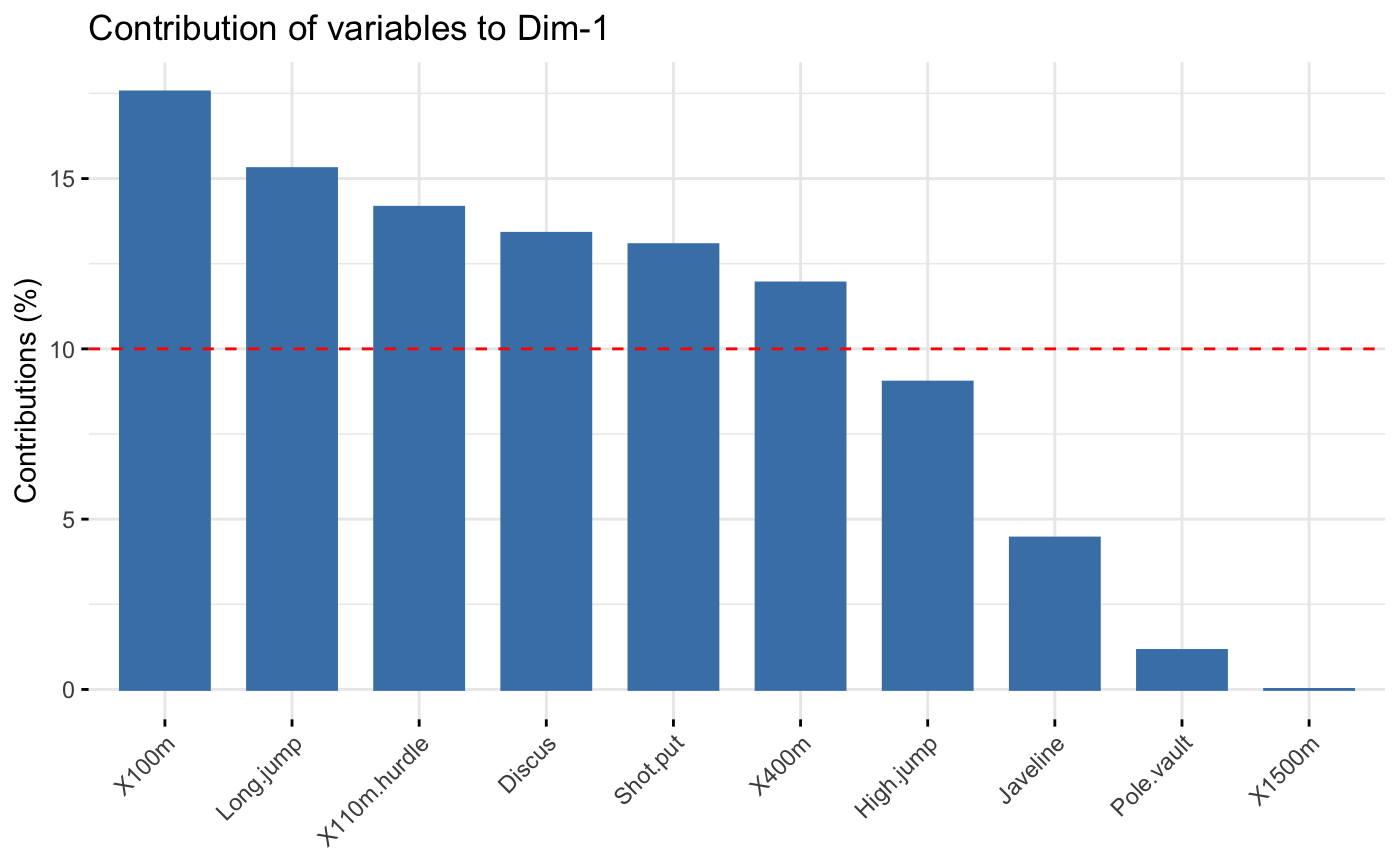
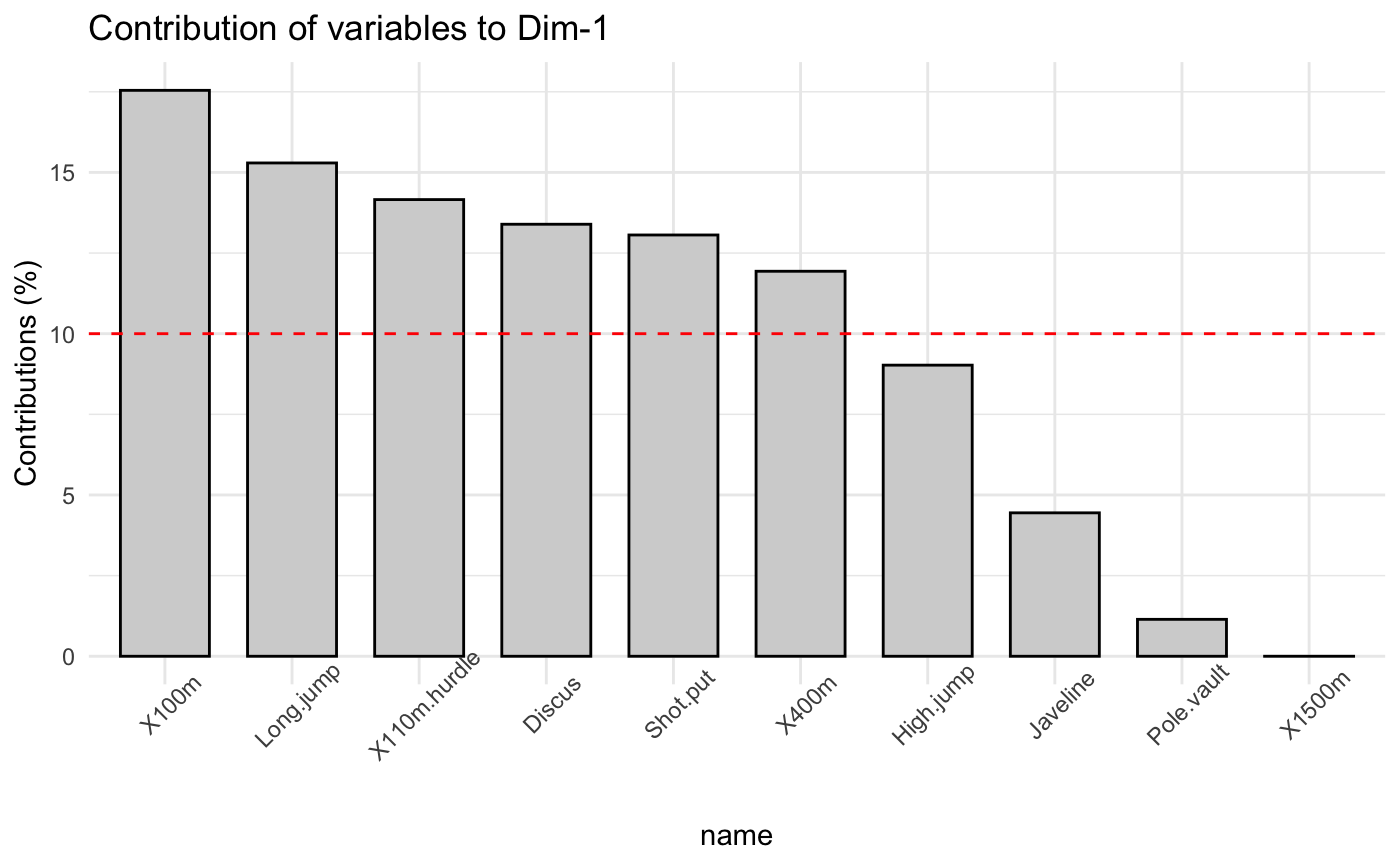
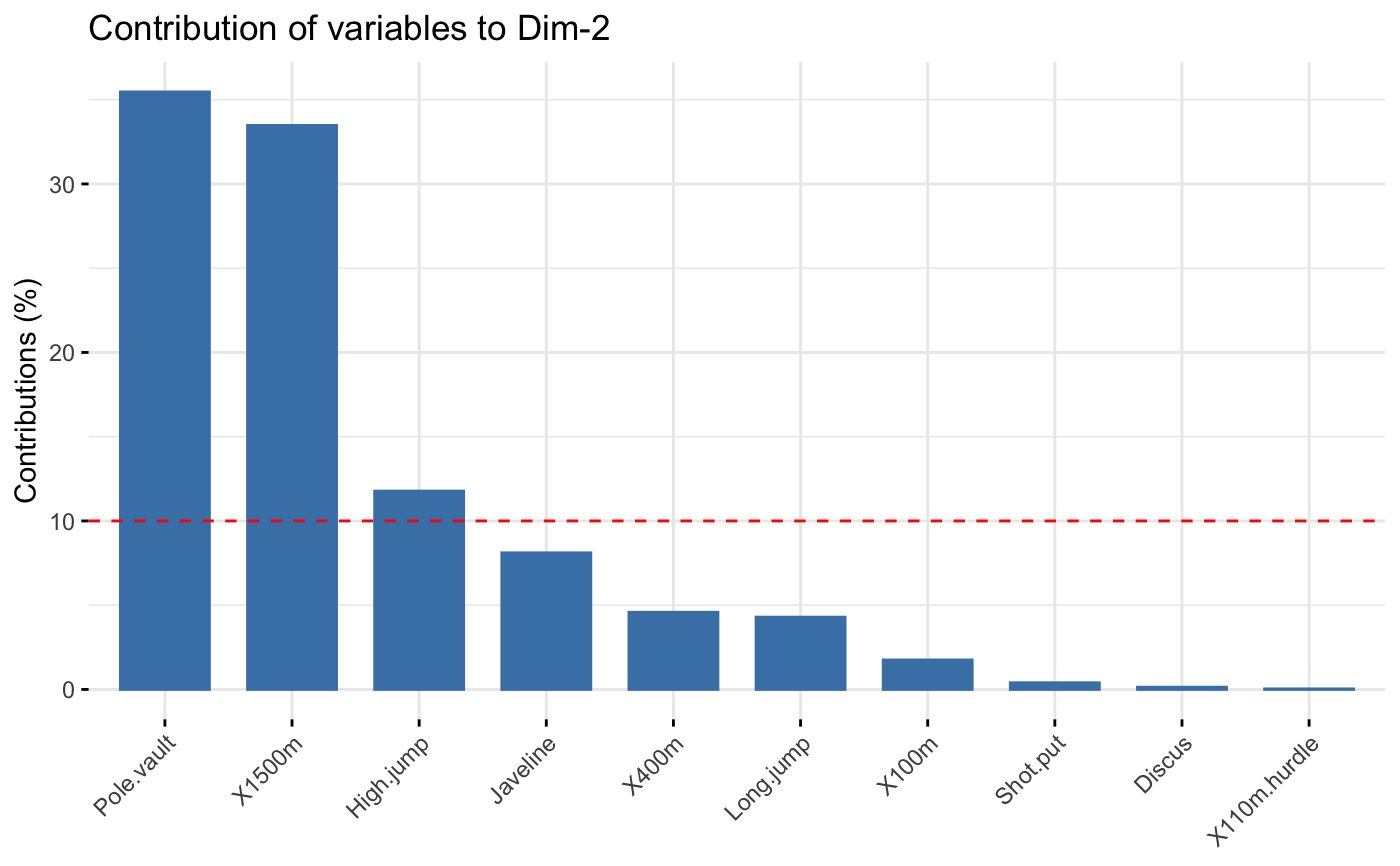
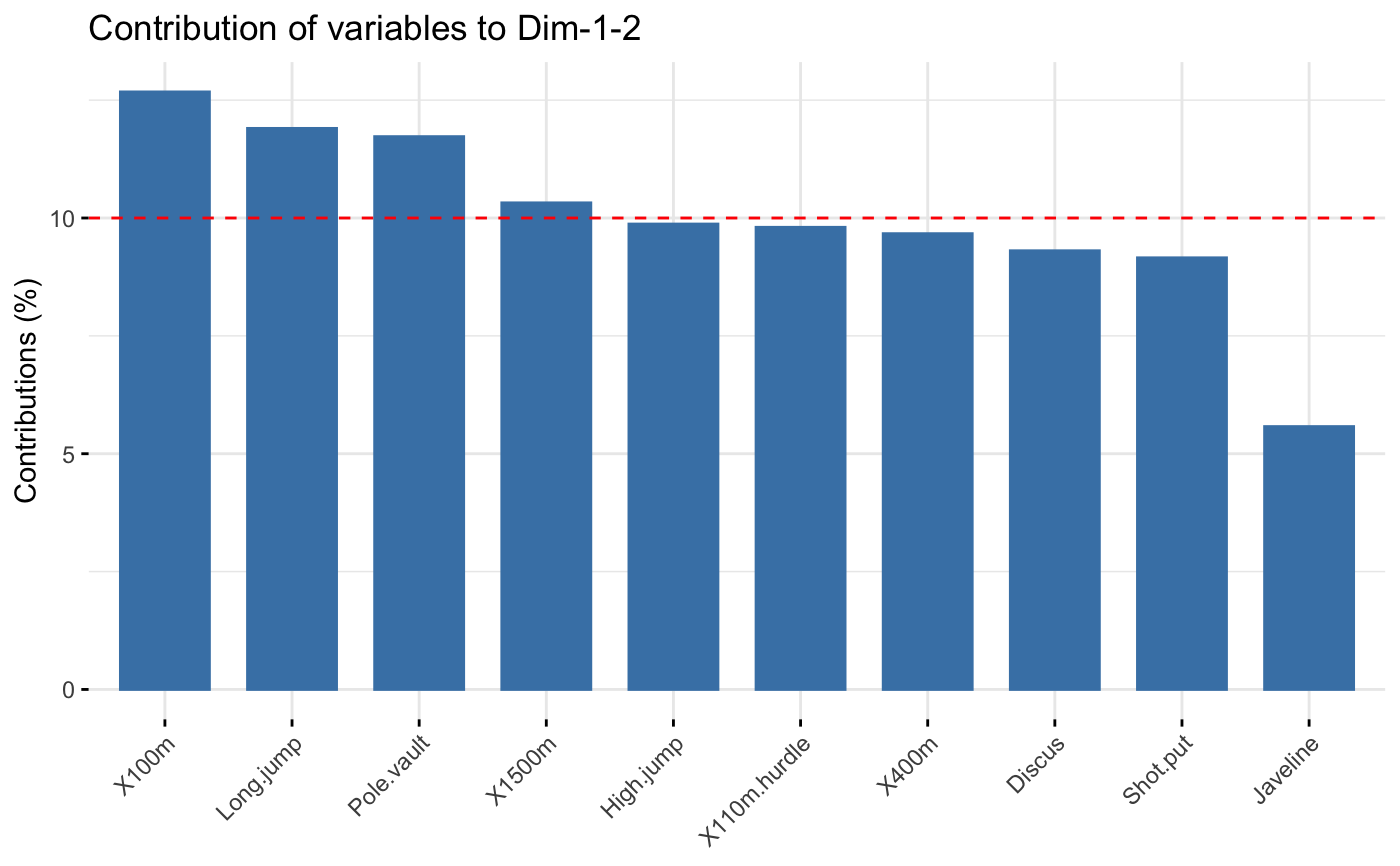
The function fviz_contrib() creates a barplot of row/column contributions.
A reference dashed line is also shown on the barplot. This reference line
corresponds to the expected value if the contribution where uniform.
For a given dimension, any row/column with a contribution above the reference line could be
considered as important in contributing to the dimension.
Functions
fviz_pca_contrib: deprecated function. Use fviz_contrib()
References
http://www.sthda.com/english/
Examples
# \donttest{ # Principal component analysis # ++++++++++++++++++++++++++ data(decathlon2) decathlon2.active <- decathlon2[1:23, 1:10] res.pca <- prcomp(decathlon2.active, scale = TRUE) # variable contributions on axis 1 fviz_contrib(res.pca, choice="var", axes = 1, top = 10 )# Change theme and color fviz_contrib(res.pca, choice="var", axes = 1, fill = "lightgray", color = "black") + theme_minimal() + theme(axis.text.x = element_text(angle=45))# Variable contributions on axis 2 fviz_contrib(res.pca, choice="var", axes = 2)# Variable contributions on axes 1 + 2 fviz_contrib(res.pca, choice="var", axes = 1:2)# Contributions of individuals on axis 1 fviz_contrib(res.pca, choice="ind", axes = 1)if (FALSE) { # Correspondence Analysis # ++++++++++++++++++++++++++ # Install and load FactoMineR to compute CA # install.packages("FactoMineR") library("FactoMineR") data("housetasks") res.ca <- CA(housetasks, graph = FALSE) # Visualize row contributions on axes 1 fviz_contrib(res.ca, choice ="row", axes = 1) # Visualize column contributions on axes 1 fviz_contrib(res.ca, choice ="col", axes = 1) # Multiple Correspondence Analysis # +++++++++++++++++++++++++++++++++ library(FactoMineR) data(poison) res.mca <- MCA(poison, quanti.sup = 1:2, quali.sup = 3:4, graph=FALSE) # Visualize individual contributions on axes 1 fviz_contrib(res.mca, choice ="ind", axes = 1) # Visualize variable categorie contributions on axes 1 fviz_contrib(res.mca, choice ="var", axes = 1) # Multiple Factor Analysis # ++++++++++++++++++++++++ library(FactoMineR) data(poison) res.mfa <- MFA(poison, group=c(2,2,5,6), type=c("s","n","n","n"), name.group=c("desc","desc2","symptom","eat"), num.group.sup=1:2, graph=FALSE) # Visualize individual contributions on axes 1 fviz_contrib(res.mfa, choice ="ind", axes = 1, top = 20) # Visualize catecorical variable categorie contributions on axes 1 fviz_contrib(res.mfa, choice ="quali.var", axes = 1) } # }